Difference between revisions of "Porichthys notatus"
From Ucsbgalaxy
| Line 5: | Line 5: | ||
# Express luciferase protein and assay activity | # Express luciferase protein and assay activity | ||
# Identify closely related genes from Porichtys and other fish genomes. Hypothesis: luciferase is derived from digestive enzymes. | # Identify closely related genes from Porichtys and other fish genomes. Hypothesis: luciferase is derived from digestive enzymes. | ||
| + | |||
| + | ===History=== | ||
| + | # Prepared cDNA and sent to Duke for 454 sequencing. This was an early 454 run. The first cDNA was not concentrated enough, due to using nanodrop instead of Qbit. 454 run was done on new cDNA Feb 20, 2008. Data for the run is in the phylogeny database: | ||
| + | [http://macroevolution.lifesci.ucsb.edu/phylogeny/454runlist.php ] | ||
| + | |||
| + | This was our first 454 run, and it is not great. Only 42K reads. Assembly and histograms were done in Newbler 2.6 using Galaxy. The Galaxy History is public[http://knot.cnsi.ucsb.edu:8080/u/ostratodd/h/poricthys-photophore] | ||
Revision as of 00:30, 12 November 2011
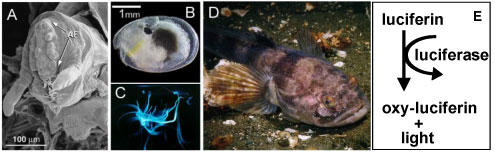
Identification of Midshipman Luciferase
Goals
- Identify midshipman (Porichthys notatus) luciferase
- Express luciferase protein and assay activity
- Identify closely related genes from Porichtys and other fish genomes. Hypothesis: luciferase is derived from digestive enzymes.
History
- Prepared cDNA and sent to Duke for 454 sequencing. This was an early 454 run. The first cDNA was not concentrated enough, due to using nanodrop instead of Qbit. 454 run was done on new cDNA Feb 20, 2008. Data for the run is in the phylogeny database:
This was our first 454 run, and it is not great. Only 42K reads. Assembly and histograms were done in Newbler 2.6 using Galaxy. The Galaxy History is public[2]